We’re extremely excited to share the Deep Graph Knowledge Embedding Library (DGL-KE), a knowledge graph (KG) embeddings library built on top of the Deep Graph Library (DGL). DGL is an easy-to-use, high-performance, scalable Python library for deep learning on graphs. You can now create embeddings for large KGs containing billions of nodes and edges two-to-five times faster than competing techniques.
For example, DGL-KE has created embeddings on top of the Drug Repurposing Knowledge Graph (DRKG) to show which drugs can be repurposed to fight COVID-19. These embeddings can be used to predict the likelihood of a drug’s ability to treat a disease or bind to a protein associated with the disease.
In this post, we focus on creating knowledge graph embeddings (KGE) using the Kensho Derived Wikimedia Dataset (KDWD). You can use those embeddings to find similar nodes and predict new relations. For example, in natural language processing (NLP) and information retrieval use cases, you can parse a new query and transform it syntactically into a triplet (subject, predicate, object). Upon adding new triplets to a KG, you can augment nodes and relations by classifying nodes and inferring relations based on the existing KG embeddings. This helps guide and find the intent for a chatbot application, for example, and provide the right FAQ or information to a customer.
Knowledge graph
A knowledge graph is a structured representation of facts, consisting of entities, relationships, and semantic descriptions purposely built for a given domain or application. They are also known as heterogenous graphs, where there are multiple entity types and relation types. The information stored in a KG is often specified in triplets, which contain three elements: head, relation, and tail ([h,r,t]). Heads and tails are also known as entities. The union of triplets is also known as statements.
KGs allow you to model your information very intuitively and expressively, giving you the ability to integrate data easily. For example, you can use Amazon Neptune to build an identity graph powering your customer 360 or Know Your Customer application commonly found in financial services. In healthcare and life sciences, where data is usually sparse, KGs can integrate and harmonize data from different silos using taxonomy and vocabularies. In e-commerce and telco, KGs are commonly used in question answering, chatbots, and recommender systems. Fore more information on using Amazon Neptune for your use case, visit the Amazon Neptune homepage.
Knowledge graph embeddings
Knowledge graph embeddings are low-dimensional representations of the entities and relations in a knowledge graph. They generalize information of the semantic and local structure for a given node.
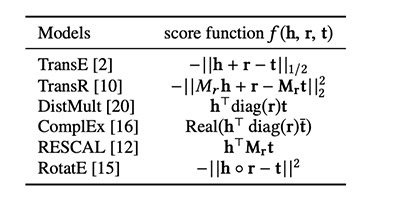
Many popular KGE models exist, such as TransE, TransR, RESCAL, DistMult, ComplEx, and RotatE.
Each model has a different score function that measures the distance of two associated entities by their relation. The general intuition is that entities connected by a relation are closed to each other, whereas the entities that aren’t connected are far apart in the vector space.
The scoring functions for the models currently supported by DGL-KE are as follows:
Wikimedia dataset
In our use case, we use the Kensho Derived Wikimedia Dataset (KDWD). You can find the notebook example and code in the DGL-KE GitHub repo.
The combination of Wikipedia and Wikidata is composed of three data layers:
- Base – Contains the English Wikipedia corpus
- Middle – Identifies which text spans are links and annotates the corpus
- Top – Connects Wikipedia links to items in the Wikidata KG
The following diagram illustrates these data layers.

The KDWD contains the following:
- 2,315,761,359 tokens
- 121,835,453 page links
- 5,343,564 Wikipedia pages
- 51,450,317 Wikidata items
- 141,206,854 Wikidata statements
The following code is an example of the entity.txt file:
ID Label Description
1 Universe totality of space and all contents
2 Earth third planet from the Sun in the Solar System
3 life matter capable of extracting energy from the environment for replicationBefore you can create your embeddings, you need to pre-process the data. DGL-KE gives you the ability to compute embeddings using two formats.
In raw user-defined knowledge graphs, you provide the triplets; the entities and relations can be arbitrary strings. The dataloader automatically generates the ID mappings.
The following table from Train User-Defined Knowledge Graphs shows an example of triplets.
| train.tsv | ||
| Beijing | is_capital_of | China |
| London | is_capital_of | UK |
| UK | located_at | Europe |
| … |
In user-defined knowledge graphs, you also provide the ID mapping for entities and relations (the triplets should only contain these IDs). The IDs start from 0 and are continuous. The following table from Train User-Defined Knowledge Graphs shows an example of mapping and triplets files.
| entities.dict | relation.dict | train.tsv |
| Beijing 0 | is_capital_of 0 | 0 0 2 |
| London 1 | located_at 1 | 1 0 3 |
| China 2 | 3 1 4 | |
| UK 3 | ||
| Europe 4 |
For more information, see DGL-KE Command Lines.
Although the dataset KDWD provides dictionaries, we can’t use it for our use case because the index doesn’t start with 0 and the index values aren’t continuous. We preprocess our data and use the raw format to generate our embeddings. After merging and cleaning the data, we end up with a KG with the following properties:
- 39,569,815 entities
- 1,213 relations
- Approximately 120 million statements
The following code is an example of the triplets.
Head Relation Tail
Eiksteinen located in the administrative… Rogaland
Trivellona marlowi instance of taxon
Acta Numerica main subject mathematical analysis
Günther Neukirchner given name Günther
Ruth Pointer given name Ruth
DGL-KE has many different training modes. CPU, GPU, mix-CPU-GPU mode, and distributed training are all supported options, depending on your dataset and training requirements. For our use case, we use mix mode to generate our embeddings. If you can contain all the data in GPU memory, GPU is the preferred method. Because we’re training on a large KG, we use mix mode to get a larger pool of CPU- and GPU-based memory and still benefit from GPU for accelerated training.
We create our embeddings with the dgl-ke command line. See the following code:
!DGLBACKEND=pytorch dglke_train
--model_name TransE_l2
--batch_size 1000
--neg_sample_size 200
--hidden_dim 400
--gamma 19.9
--lr 0.25
--max_step 24000
--log_interval 100
--batch_size_eval 16
-adv
--regularization_coef 1.00E-09
--test
--gpu 0 1 2 3
--mix_cpu_gpu
--save_path ./wikimedia
--data_path ./data/wikimedia/
--format raw_udd_hrt
--data_files train.txt valid.txt test.txt
--neg_sample_size_eval 10000
For more information about the DGL-KE arguments, see the DGL-KE website.
We trained our KG of about 40 million entities and 1,200 relations and approximately 120 million statements on a p3.8xl in about 7 minutes.
We evaluate our model by entering the following code:
!DGLBACKEND=pytorch dglke_eval --dataset wikimedia --model_name TransE_l2
--neg_sample_size 200 --hidden_dim 400 --gamma 19.9
--batch_size_eval 16 --gpu 0 1 2 3 --model_path ./wikimedia/TransE_l2_wikimedia_0/
--data_path ./data/wikimedia/ --format raw_udd_hrt --data_files train.txt valid.txt test.txt --neg_sample_size_eval 10000 --no_eval_filter
The following code is the output:
-------------- Test result --------------
Test average MRR: 0.4159753346227368
Test average MR: 1001.1689418833716
Test average HITS@1: 0.3540242971873324
Test average HITS@3: 0.45541123141672746
Test average HITS@10: 0.5213350742247359
-----------------------------------------DGL-KE allows you to perform KG downstream tasks by using any combinations of [h,r,t].
In the following example, we find similar node entities for two people by creating a head.list file with the following entries:
head.list
Jeff Bezos
Barack ObamaDGL-KE provides functions to perform offline inference on entities and relations.
To find similar node entities from our head.list, we enter the following code:
!DGLBACKEND=pytorch dglke_emb_sim
--format 'l_*' --data_files /home/ec2-user/SageMaker/DGL-Neptune/notebooks/data/wikimedia/head.list
--mfile ./data/wikimedia/entities.tsv
--emb_file ./wikimedia/TransE_l2_wikimedia_1/wikimedia_TransE_l2_entity.npy
--raw_data --gpu 0
--exec_mode 'batch_left'
--sim_func 'cosine' --topK 5
The following code is the output:
resulst.tsv
head tail score
Jeff Bezos Jeff Bezos 1.0
Jeff Bezos Aga Khan IV 0.8602205514907837
Jeff Bezos Alisher Usmanov 0.8584005236625671
Jeff Bezos Klaus Tschira 0.8512368202209473
Jeff Bezos Bill Gates 0.8441287875175476
Barack Obama Barack Obama 1.0
Barack Obama Donald Trump 0.9529082179069519
Barack Obama George W. Bush 0.9426612854003906
Barack Obama Harry S. Truman 0.9414601922035217
Barack Obama Ronald Reagan 0.9393566250801086Interestingly, all the nodes similar to Jeff Bezos describe tech tycoons. Barack Obama’s similar nodes show former and current presidents of the US.
Conclusion
Graphs can be found in many domains, such as chemistry, biology, financial services, and social networks, and allow us to represent complex concepts intuitively using entities and relations. Graphs can be homogeneous or heterogeneous, where you have many types of entities and relations.
Knowledge graph embeddings give you powerful methods to encode semantic and local structure information for a given node, and you can also use them as input for machine learning and deep learning models. DGL-KE supports popular embedding models and allows you to compute those embeddings on CPU or GPU at scale two-to-five times faster than other techniques.
We’re excited to see how you use graph embeddings on your existing KGs or new machine learning problems. For more information about the library, see the DGL-KE GitHub repo. For instructions on using Wikimedia KG embeddings in your KG, see the DGL-KE notebook example.
About the Authors
 Phi Nguyen is a solution architect at AWS helping customers with their cloud journey with a special focus on data lake, analytics, semantics technologies and machine learning. In his spare time, you can find him biking to work, coaching his son’s soccer team or enjoying nature walk with his family.
Phi Nguyen is a solution architect at AWS helping customers with their cloud journey with a special focus on data lake, analytics, semantics technologies and machine learning. In his spare time, you can find him biking to work, coaching his son’s soccer team or enjoying nature walk with his family.
 Xiang Song is an Applied Scientist with the AWS Shanghai AI Lab. He got his Bachelor’s degree in Software Engineer and Ph.D’s in Operating System and Architecture from Fudan University. His research interests include building machine learning systems and graph neural network for real world applications.
Xiang Song is an Applied Scientist with the AWS Shanghai AI Lab. He got his Bachelor’s degree in Software Engineer and Ph.D’s in Operating System and Architecture from Fudan University. His research interests include building machine learning systems and graph neural network for real world applications.
