Posted by Tim Blakely, Software Engineer and Michał Januszewski, Research Scientist, Connectomics at Google
In January 2020 we released the fly “hemibrain” connectome — an online database providing the morphological structure and synaptic connectivity of roughly half of the brain of a fruit fly (Drosophila melanogaster). This database and its supporting visualization has reframed the way that neural circuits are studied and understood in the fly brain. While the fruit fly brain is small enough to attain a relatively complete map using modern mapping techniques, the insights gained are, at best, only partially informative to understanding the most interesting object in neuroscience — the human brain.
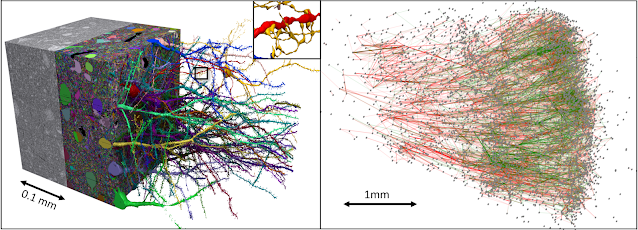
Today, in collaboration with the Lichtman Laboratory at Harvard University, we are releasing the “H01” dataset, a 1.4 petabyte rendering of a small sample of human brain tissue, along with a companion paper, “A connectomic study of a petascale fragment of human cerebral cortex.” The H01 sample was imaged at 4nm-resolution by serial section electron microscopy, reconstructed and annotated by automated computational techniques, and analyzed for preliminary insights into the structure of the human cortex. The dataset comprises imaging data that covers roughly one cubic millimeter of brain tissue, and includes tens of thousands of reconstructed neurons, millions of neuron fragments, 130 million annotated synapses, 104 proofread cells, and many additional subcellular annotations and structures — all easily accessible with the Neuroglancer browser interface. H01 is thus far the largest sample of brain tissue imaged and reconstructed in this level of detail, in any species, and the first large-scale study of synaptic connectivity in the human cortex that spans multiple cell types across all layers of the cortex. The primary goals of this project are to produce a novel resource for studying the human brain and to improve and scale the underlying connectomics technologies.
What is the Human Cortex?
The cerebral cortex is the thin surface layer of the brain found in vertebrate animals that has evolved most recently, showing the greatest variation in size among different mammals (it is especially large in humans). Each part of the cerebral cortex is six layered (e.g., L2), with different kinds of nerve cells (e.g., spiny stellate) in each layer. The cerebral cortex plays a crucial role in most higher level cognitive functions, such as thinking, memory, planning, perception, language, and attention. Although there has been some progress in understanding the macroscopic organization of this very complicated tissue, its organization at the level of individual nerve cells and their interconnecting synapses is largely unknown.
Human Brain Connectomics: From Surgical Biopsy to a 3D Database
Mapping the structure of the brain at the resolution of individual synapses requires high-resolution microscopy techniques that can image biochemically stabilized (fixed) tissue. We collaborated with brain surgeons at Massachusetts General Hospital in Boston (MGH) who sometimes remove pieces of normal human cerebral cortex when performing a surgery to cure epilepsy in order to gain access to a site in the deeper brain where an epileptic seizure is being initiated. Patients anonymously donated this tissue, which is normally discarded, to our colleagues in the Lichtman lab. The Harvard researchers cut the tissue into ~5300 individual 30 nanometer sections using an automated tape collecting ultra-microtome, mounted those sections onto silicon wafers, and then imaged the brain tissue at 4 nm resolution in a customized 61-beam parallelized scanning electron microscope for rapid image acquisition.
Imaging the ~5300 physical sections produced 225 million individual 2D images. Our team then computationally stitched and aligned this data to produce a single 3D volume. While the quality of the data was generally excellent, these alignment pipelines had to robustly handle a number of challenges, including imaging artifacts, missing sections, variation in microscope parameters, and physical stretching and compression of the tissue. Once aligned, a multiscale flood-filling network pipeline was applied (using thousands of Google Cloud TPUs) to produce a 3D segmentation of each individual cell in the tissue. Additional machine learning pipelines were applied to identify and characterize 130 million synapses, classify each 3D fragment into various “subcompartments” (e.g., axon, dendrite, or cell body), and identify other structures of interest such as myelin and cilia. Automated reconstruction results were imperfect, so manual efforts were used to “proofread” roughly one hundred cells in the data. Over time, we expect to add additional cells to this verified set through additional manual efforts and further advances in automation.
| The H01 volume: roughly one cubic millimeter of human brain tissue captured in 1.4 petabytes of images. |
The imaging data, reconstruction results, and annotations are viewable through an interactive web-based 3D visualization interface, called Neuroglancer, that was originally developed to visualize the fruit fly brain. Neuroglancer is available as open-source software, and widely used in the broader connectomics community. Several new features were introduced to support analysis of the H01 dataset, in particular support for searching for specific neurons in the dataset based on their type or other properties.
 |
 |
 |
||
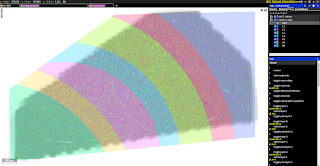
| The volume spans all six cortical layers | Highlighting Layer 2 interneurons | Excitatory and inhibitory incoming synapses |
 |
 |
 |
||
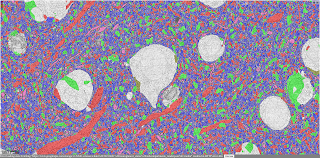
| Neuronal subcompartments classified | A Chandelier cell and some of the Pyramidal neurons it inhibits | Blood vessels traced throughout the volume |
 |
 |
 |
||
| Serial contact between a pair of neurons | An axon with elaborate whorl structures | A neuron with unusual propensity for self-contact (Credit: Rachael Han) |
| The Neuroglancer interface to the H01 volume and annotations. The user can select specific cells on the basis of their layer and type, can view incoming and outgoing synapses for the cell, and much more. (Click on the images above to take you to the Neuroglancer view shown.) |
Analysis of the Human Cortex
In a companion preprint, we show how H01 has already been used to study several interesting aspects of the organization of the human cortex. In particular, new cell types have been discovered, as well as the presence of “outlier” axonal inputs, which establish powerful synaptic connections with target dendrites. While these findings are a promising start, the vastness of the H01 dataset will provide a basis for many years of further study by researchers interested in the human cortex.
In order to accelerate the analysis of H01, we also provide embeddings of the H01 data that were generated by a neural network trained using a variant of the SimCLR self-supervised learning technique. These embeddings provide highly informative representations of local parts of the dataset that can be used to rapidly annotate new structures and develop new ways of clustering and categorizing brain structures according to purely data-driven criteria. We trained these embeddings using Google Cloud TPU pods and then performed inference at roughly four billion data locations spread throughout the volume.
Managing Dataset Size with Improved Compression
H01 is a petabyte-scale dataset, but is only one-millionth the volume of an entire human brain. Serious technical challenges remain in scaling up synapse-level brain mapping to an entire mouse brain (500x bigger than H01), let alone an entire human brain. One of these challenges is data storage: a mouse brain could generate an exabyte worth of data, which is costly to store. To address this, we are today also releasing a paper, “Denoising-based Image Compression for Connectomics”, that details how a machine learning-based denoising strategy can be used to compress data, such as H01, at least 17-fold (dashed line in the figure below), with negligible loss of accuracy in the automated reconstruction.
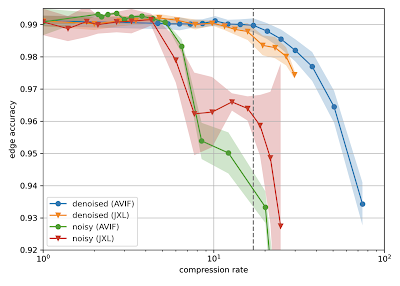 |
| Reconstruction quality of noisy and denoised images as a function of compression rate for JPEG XL (JXL) and AV Image Format (AVIF) codecs. Points and lines show the means, and the shaded area covers ±1 standard deviation around the mean. |
Random variations in the electron microscopy imaging process lead to image noise that is difficult to compress even in principle, as the noise lacks spatial correlations or other structure that could be described with fewer bytes. Therefore we acquired images of the same piece of tissue in both a “fast” acquisition regime (resulting in high amounts of noise) and a “slow” acquisition regime (resulting in low amounts of noise) and then trained a neural network to infer “slow” scans from “fast” scans. Standard image compression codecs were then able to (lossily) compress the “virtual” slow scans with fewer artifacts compared to the raw data. We believe this advance has the potential to significantly mitigate the costs associated with future large scale connectomics projects.
Next Steps
But storage is not the only problem. The sheer size of future data sets will require developing new strategies for researchers to organize and access the rich information inherent in connectomic data. These are challenges that will require new modes of interaction between humans and the brain mapping data that will be forthcoming.