
The combination of the environment an individual experiences and their genetic predispositions determines the majority of their risk for various diseases. Large national efforts, such as the UK Biobank, have created large, public resources to better understand the links between environment, genetics, and disease. This has the potential to help individuals better understand how to stay healthy, clinicians to treat illnesses, and scientists to develop new medicines.
One challenge in this process is how we make sense of the vast amount of clinical measurements — the UK Biobank has many petabytes of imaging, metabolic tests, and medical records spanning 500,000 individuals. To best use this data, we need to be able to represent the information present as succinct, informative labels about meaningful diseases and traits, a process called phenotyping. That is where we can use the ability of ML models to pick up on subtle intricate patterns in large amounts of data.
We’ve previously demonstrated the ability to use ML models to quickly phenotype at scale for retinal diseases. Nonetheless, these models were trained using labels from clinician judgment, and access to clinical-grade labels is a limiting factor due to the time and expense needed to create them.
In “Inference of chronic obstructive pulmonary disease with deep learning on raw spirograms identifies new genetic loci and improves risk models”, published in Nature Genetics, we’re excited to highlight a method for training accurate ML models for genetic discovery of diseases, even when using noisy and unreliable labels. We demonstrate the ability to train ML models that can phenotype directly from raw clinical measurement and unreliable medical record information. This reduced reliance on medical domain experts for labeling greatly expands the range of applications for our technique to a panoply of diseases and has the potential to improve their prevention, diagnosis, and treatment. We showcase this method with ML models that can better characterize lung function and chronic obstructive pulmonary disease (COPD). Additionally, we show the usefulness of these models by demonstrating a better ability to identify genetic variants associated with COPD, improved understanding of the biology behind the disease, and successful prediction of outcomes associated with COPD.
ML for deeper understanding of exhalation
For this demonstration, we focused on COPD, the third leading cause of worldwide death in 2019, in which airway inflammation and impeded airflow can progressively reduce lung function. Lung function for COPD and other diseases is measured by recording an individual’s exhalation volume over time (the record is called a spirogram; see an example below). Although there are guidelines (called GOLD) for determining COPD status from exhalation, these use only a few, specific data points in the curve and apply fixed thresholds to those values. Much of the rich data from these spirograms is discarded in this analysis of lung function.
We reasoned that ML models trained to classify spirograms would be able to use the rich data present more completely and result in more accurate and comprehensive measures of lung function and disease, similar to what we have seen in other classification tasks like mammography or histology. We trained ML models to predict whether an individual has COPD using the full spirograms as inputs.
The common method of training models for this problem, supervised learning, requires samples to be associated with labels. Determining those labels can require the effort of very time-constrained experts. For this work, to show that we do not necessarily need medically graded labels, we decided to use a variety of widely available sources of medical record information to create those labels without medical expert review. These labels are less reliable and noisy for two reasons. First, there are gaps in the medical records of individuals because they use multiple health services. Second, COPD is often undiagnosed, meaning many with the disease will not be labeled as having it even if we compile the complete medical records. Nonetheless, we trained a model to predict these noisy labels from the spirogram curves and treat the model predictions as a quantitative COPD liability or risk score.
 |
| Noisy COPD status labels were derived using various medical record sources (clinical data). A COPD liability model is then trained to predict COPD status from raw flow-volume spirograms. |
Predicting COPD outcomes
We then investigated whether the risk scores produced by our model could better predict a variety of binary COPD outcomes (for example, an individual’s COPD status, whether they were hospitalized for COPD or died from it). For comparison, we benchmarked the model relative to expert-defined measurements required to diagnose COPD, specifically FEV1/FVC, which compares specific points on the spirogram curve with a simple mathematical ratio. We observed an improvement in the ability to predict these outcomes as seen in the precision-recall curves below.
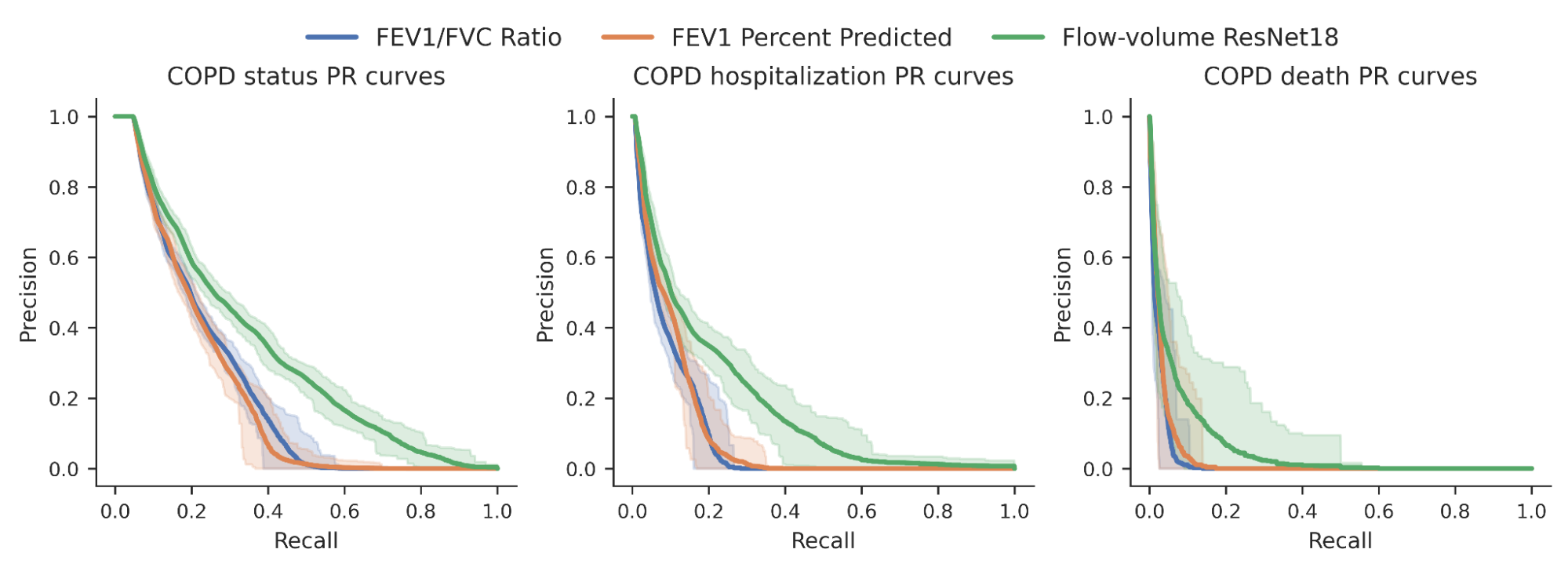 |
| Precision-recall curves for COPD status and outcomes for our ML model (green) compared to traditional measures. Confidence intervals are shown by lighter shading. |
We also observed that separating populations by their COPD model score was predictive of all-cause mortality. This plot suggests that individuals with higher COPD risk are more likely to die earlier from any causes and the risk probably has implications beyond just COPD.
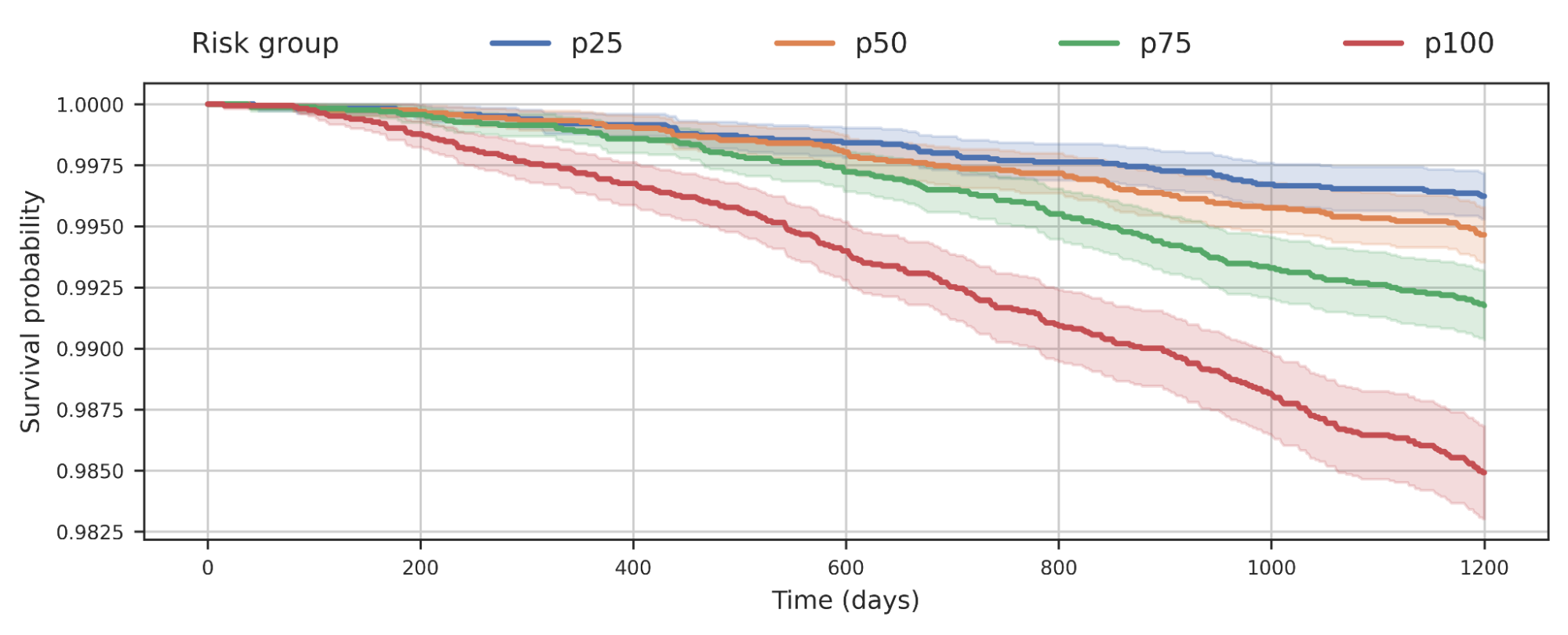 |
| Survival analysis of a cohort of UK Biobank individuals stratified by their COPD model’s predicted risk quartile. The decrease of the curve indicates individuals in the cohort dying over time. For example, p100 represents the 25% of the cohort with greatest predicted risk, while p50 represents the 2nd quartile. |
Identifying the genetic links with COPD
Since the goal of large scale biobanks is to bring together large amounts of both phenotype and genetic data, we also performed a test called a genome-wide association study (GWAS) to identify the genetic links with COPD and genetic predisposition. A GWAS measures the strength of the statistical association between a given genetic variant — a change in a specific position of DNA — and the observations (e.g., COPD) across a cohort of cases and controls. Genetic associations discovered in this manner can inform drug development that modifies the activity or products of a gene, as well as expand our understanding of the biology for a disease.
We showed with our ML-phenotyping method that not only do we rediscover almost all known COPD variants found by manual phenotyping, but we also find many novel genetic variants significantly associated with COPD. In addition, we see good agreement on the effect sizes for the variants discovered by both our ML approach and the manual one (R2=0.93), which provides strong evidence for validity of the newly found variants.
Finally, our collaborators at Harvard Medical School and Brigham and Women’s Hospital further examined the plausibility of these findings by providing insights into the possible biological role of the novel variants in development and progression of COPD (you can see more discussion on these insights in the paper).
Conclusion
We demonstrated that our earlier methods for phenotyping with ML can be expanded to a wide range of diseases and can provide novel and valuable insights. We made two key observations by using this to predict COPD from spirograms and discovering new genetic insights. First, domain knowledge was not necessary to make predictions from raw medical data. Interestingly, we showed the raw medical data is probably underutilized and the ML model can find patterns in it that are not captured by expert-defined measurements. Second, we do not need medically graded labels; instead, noisy labels defined from widely available medical records can be used to generate clinically predictive and genetically informative risk scores. We hope that this work will broadly expand the ability of the field to use noisy labels and will improve our collective understanding of lung function and disease.
Acknowledgments
This work is the combined output of multiple contributors and institutions. We thank all contributors: Justin Cosentino, Babak Alipanahi, Zachary R. McCaw, Cory Y. McLean, Farhad Hormozdiari (Google), Davin Hill (Northeastern University), Tae-Hwi Schwantes-An and Dongbing Lai (Indiana University), Brian D. Hobbs and Michael H. Cho (Brigham and Women’s Hospital, and Harvard Medical School). We also thank Ted Yun and Nick Furlotte for reviewing the manuscript, Greg Corrado and Shravya Shetty for support, and Howard Yang, Kavita Kulkarni, and Tammi Huynh for helping with publication logistics.


