Medical data is highly contextual and heavily multi-modal, in which each data silo is treated separately. To bridge different data, a knowledge graph-based approach integrates data across domains and helps represent the complex representation of scientific knowledge more naturally. For example, three components of major electronic health records (EHR) are diagnosis codes, primary notes, and specific medications. Because these are represented in different data silos, secondary use of these documents for accurately identifying patients with a specific observable trait is a crucial challenge. By connecting those different sources, subject matter experts have a richer pool of data to understand how different concepts such as diseases and symptoms interact with one another and help conduct their research. This ultimately helps healthcare and life sciences researchers and practitioners create better insights from the data for a variety of use cases, such as drug discovery and personalized treatments.
In this post, we use Amazon HealthLake to export EHR data in the Fast Healthcare Interoperability Resources (FHIR) data format. We then build a knowledge graph based on key entities extracted and harmonized from the medical data. Amazon HealthLake also extracts and transforms unstructured medical data, such as medical notes, so it can be searched and analyzed. Together with Amazon Kendra and Amazon Neptune, we allow domain experts to ask a natural language question, surface the results and relevant documents, and show connected key entities such as treatments, inferred ICD-10 codes, medications, and more across records and documents. This allows for easy analysis of co-occurrence of key entities, co-morbidities analysis, and patient cohort analysis in an integrated solution. Combining effective search capabilities and data mining through graph networks reduces time and cost for users to find relevant information around patients and improve knowledge serviceability surrounding EHRs. The code base for this post is available on the GitHub repo.
Solution overview
In this post, we use the output from Amazon HealthLake for two purposes.
First, we index EHRs into Amazon Kendra for semantic and accurate document ranking out of patient notes, which help improve physician efficiency identifying patient notes and compare it with other patients sharing similar characteristics. This shifts from using a lexical search to a semantic search that introduces context around the query, which results in better search output (see the following screenshot).

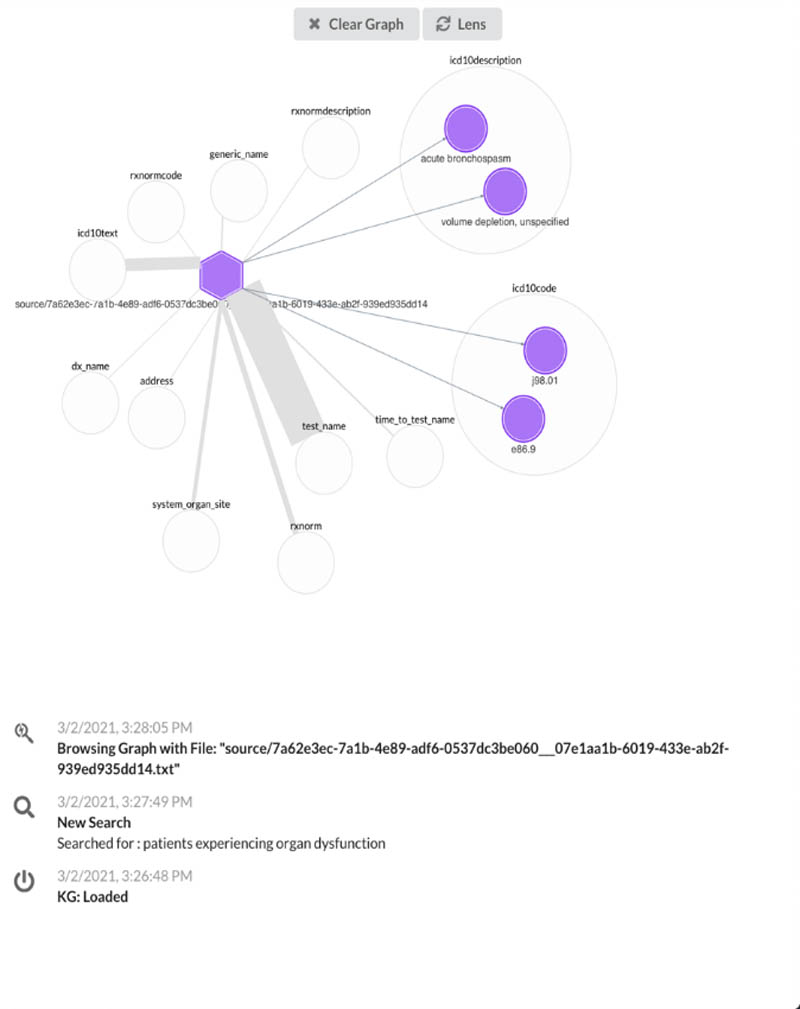
Second, we use Neptune to build knowledge graph applications for users to view metadata associated with patient notes in a more simple and normalized view, which allows us to highlight the important characteristics stemming from a document (see the following screenshot).
The following diagram illustrates our architecture.

The steps to implement the solution are as follows:
- Create and export Amazon HealthLake data.
- Extract patient visit notes and metadata.
- Load patient notes data into Amazon Kendra.
- Load the data into Neptune.
- Set up the backend and front end to run the web app.
Create and export Amazon HealthLake data
As a first step, create a data store using Amazon HealthLake either via the Amazon HealthLake console or the AWS Command Line Interface (AWS CLI). For this post, we focus on the AWS CLI approach.
- We use AWS Cloud9 to create a data store with the following code, replacing <<your data store name >> with a unique name:
aws healthlake create-fhir-datastore --region us-east-1 --datastore-type-version R4 --preload-data-config PreloadDataType="SYNTHEA" --datastore-name "<<your_data_store_name>>"The preceding code uses a preloaded dataset from Synthea, which is supported in FHIR version R4, to explore how to use Amazon HealthLake output. Running the code produces a response similar to the following code, and this step takes a few minutes to complete (approximately 30 minutes at the time of writing):
{
"DatastoreEndpoint": "https://healthlake.us-east-1.amazonaws.com/datastore/<<your_data_store_id>>/r4/",
"DatastoreArn": "arn:aws:healthlake:us-east-1:<<your_AWS_account_number>>:datastore/fhir/<<your_data_store_id>>",
"DatastoreStatus": "CREATING",
"DatastoreId": "<<your_data_store_id>>"
}
You can check the status of completion either on the Amazon HealthLake console or in the AWS Cloud9 environment.
- To check the status in AWS Cloud9, use the following code to check the status and wait until
DatastoreStatuschanges fromCREATINGtoACTIVE:
aws healthlake describe-fhir-datastore --datastore-id "<<your_data_store_id>>" --region us-east-1- When the status changes to
ACTIVE, get the role ARN from theHEALTHLAKE-KNOWLEDGE-ANALYZER-IAMROLEstack in AWS CloudFormation, associated with the physical IDAmazonHealthLake-Export-us-east-1-HealthDataAccessRole, and copy the ARN in the linked page. - In AWS Cloud9, use the following code to export the data from Amazon HealthLake to the Amazon Simple Storage Service (Amazon S3) bucket generated from AWS Cloud Development Kit (AWS CDK) and note the
job-idoutput:
aws healthlake start-fhir-export-job --output-data-config S3Uri="s3://hl-synthea-export-<<your_AWS_account_number>>/export-$(date +"%d-%m-%y")" --datastore-id <<your_data_store_id>> --data-access-role-arn arn:aws:iam::<<your_AWS_account_number>>:role/AmazonHealthLake-Export-us-east-1-HealthKnoMaDataAccessRole- Verify that the export job is complete using the following code with the
job-idobtained from the last code you ran. (when the export is complete,JobStatusin the output statesCOMPLETED):
aws healthlake describe-fhir-export-job --datastore-id <<your_data_store_id>> --job-id <<your_job_id>>Extract patient visit notes and metadata
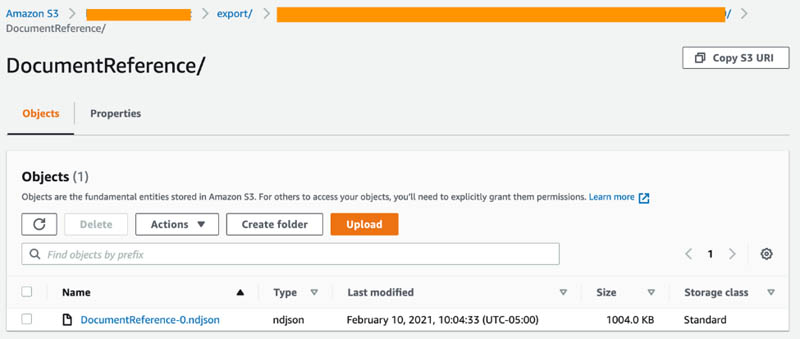
The next step involves decoding patient visits to obtain the raw texts. We will import the following file DocumentReference-0.ndjson (shown in the following screenshot of S3) from the Amazon HealthLake export step we previously completed into the CDK deployed Amazon SageMaker notebook instance. First, save the notebook provided from the Github repo into the SageMaker instance. Then, run the notebook to automatically locate and import the DocumentReference-0.ndjson files from S3.
For this step, use the resourced SageMaker to quickly run the notebook. The first part of the notebook creates a text file that contains notes from each patient’s visit and is saved to an Amazon S3 location. Because multiple visits could exist for a single patient, a unique identification combines the patient unique ID and the visit ID. These patients’ notes are used to perform semantic search against using Amazon Kendra.
The next step in the notebook involves creating triples based on the automatically extracted metadata. By creating and saving the metadata in an Amazon S3 location, an AWS Lambda function gets triggered to generate the triples surrounding the patient visit notes.
Load patient notes data into Amazon Kendra
The text files that are uploaded in the source path of the S3 bucket need to be crawled and indexed. For this post, a developer edition is created during the AWS CDK deployment, so the index is created to connect the raw patient notes.
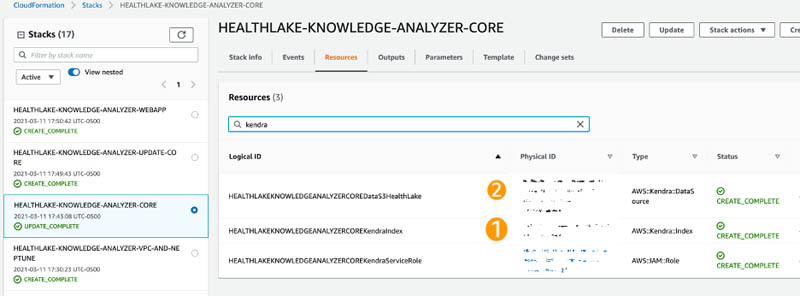
- On the AWS CloudFormation console under the HEALTHLAKE-KNOWLEDGE-ANALYZER-CORE stack, search for kendra on the Resources tab and take note of the index ID and data source ID (copy the first part of the physical ID before the pipe ( | )).
- Back in AWS Cloud9, run the following command to synchronize the patient notes in Amazon S3 to Amazon Kendra:
aws kendra start-data-source-sync-job --id <<data_source_id_2nd_circle>> --index-id <<index_id_1st_ circle>>- You can verify when the sync status is complete by running the following command:
aws kendra describe-data-source --id <<data_source_id_2nd_circle>> --index-id <<index_id_1st_circle>>Because the ingested data is very small, it should immediately show that Status is ACTIVE upon running the preceding command.
Load the data into Neptune
In this next step, we access the Amazon Elastic Compute Cloud (Amazon EC2) instance that was spun up and load the triples from Amazon S3 into Neptune using the following code:
curl -X POST
-H 'Content-Type: application/json'
https://healthlake-knowledge-analyzer-vpc-and-neptune-neptunedbcluster.cluster-<<your_unique_id>>.us-east-1.neptune.amazonaws.com:8182/loader -d '
{
"source": "s3://<<your_Amazon_S3_bucket>>/stdized-data/neptune_triples/nquads/",
"format": "nquads",
"iamRoleArn": "arn:aws:iam::<<your_AWS_account_number>>:role/KNOWLEDGE-ANALYZER-IAMROLE-ServiceRole",
"region": "us-east-1",
"failOnError": "TRUE"
}'
Set up the backend and front end to run the web app
The preceding step should take a few seconds to complete. In the meantime, configure the EC2 instance to access the web app. Make sure to have both Python and Node installed in the instance.
- Run the following code in the terminal of the instance:
sudo iptables -t nat -I PREROUTING -p tcp --dport 80 -j REDIRECT --to-ports 3000This routes the public address to the deployed app.
- Copy the two folders titled
ka-webappandka-server-webappand upload them to a folder nameddevin the EC2 instance. - For the front end, create a screen by running the following command:
screen -S back - In this screen, change the folder to
ka-webappand runnpminstall. - After installation, go into the file
.env.developmentand place the Amazon EC2 public IPv4 address and save the file. - Run
npmstart and then detach the screen. - For the backend, create another screen by entering:
screen -S back- Change the folder to
ka-server-webappand runpip install -r requirements.txt. - When the libraries are installed, enter the following code:
./run.sh- Detach from the current screen, and using any browser, go the Amazon EC2 Public IPv4 address to access the web app.
Try searching for a patient diagnosis and choose a document link to visualize the knowledge graph of that document.
Next steps
In this post, we integrate data output from Amazon HealthLake into both a search and graph engine to semantically search relevant information and highlight important entities linked to documents. You can further expand this knowledge graph and link it to other ontologies such as MeSH and MedDRA.
Furthermore, this provides a foundation to further integrate other clinical datasets and expand this knowledge graph to build a data fabric. You can make queries on historical population data, chaining structured and language-based searches for cohort selection to correlate disease with patient outcome.
Clean up
To clean up your resources, complete the following steps:
- To delete the stacks created, enter the following commands in the order given to properly remove all resources:
$ cdk destroy HEALTHLAKE-KNOWLEDGE-ANALYZER-UPDATE-CORE
$ cdk destroy HEALTHLAKE-KNOWLEDGE-ANALYZER-WEBAPP
$ cdk destroy HEALTHLAKE-KNOWLEDGE-ANALYZER-CORE
- While the preceding commands are in progress, delete the Amazon Kendra data source that was created:
$ cdk destroy HEALTHLAKE-KNOWLEDGE-ANALYZER-VPC-AND-NEPTUNE
$ cdk destroy HEALTHLAKE-KNOWLEDGE-ANALYZER-IAMROLE
$ aws healthlake delete-fhir-datastore --datastore-id <<your_data_store_id>> - To verify it’s been deleted, check the status by running the following command:
$ aws healthlake describe-fhir-datastore --datastore-id "<<your_data_store_id>>" --region us-east-1- Check the AWS CloudFormation console to ensure that all associated stacks starting with
HEALTHLAKE-KNOWLEDGE-ANALYZERhave all been deleted successfully.
Conclusion
Amazon HealthLake provides a managed service based on the FHIR standard to allow you to build health and clinical solutions. Connecting the output of Amazon HealthLake to Amazon Kendra and Neptune gives you the ability to build a cognitive search and a health knowledge graph to power your intelligent application.
Building on top of this approach can enable researchers and front-line physicians to easily search across clinical notes and research articles by simply typing their question into a web browser. Every clinical evidence is tagged, indexed, and structured using machine learning to provide evidence-based topics on things like transmission, risk factors, therapeutics, and incubation. This particular functionality is tremendously valuable for clinicians or scientists because it allows them to quickly ask a question to validate and advance their clinical decision support or research.
Try this out on your own! Deploy this solution using Amazon HealthLake in your AWS account by deploying the example on GitHub.
About the Authors
 Prithiviraj Jothikumar, PhD, is a Data Scientist with AWS Professional Services, where he helps customers build solutions using machine learning. He enjoys watching movies and sports and spending time to meditate.
Prithiviraj Jothikumar, PhD, is a Data Scientist with AWS Professional Services, where he helps customers build solutions using machine learning. He enjoys watching movies and sports and spending time to meditate.
 Phi Nguyen is a solutions architect at AWS helping customers with their cloud journey with a special focus on data lake, analytics, semantics technologies and machine learning. In his spare time, you can find him biking to work, coaching his son’s soccer team or enjoying nature walk with his fami
Phi Nguyen is a solutions architect at AWS helping customers with their cloud journey with a special focus on data lake, analytics, semantics technologies and machine learning. In his spare time, you can find him biking to work, coaching his son’s soccer team or enjoying nature walk with his fami
 Parminder Bhatia is a science leader in the AWS Health AI, currently building deep learning algorithms for clinical domain at scale. His expertise is in machine learning and large scale text analysis techniques in low resource settings, especially in biomedical, life sciences and healthcare technologies. He enjoys playing soccer, water sports and traveling with his family.
Parminder Bhatia is a science leader in the AWS Health AI, currently building deep learning algorithms for clinical domain at scale. His expertise is in machine learning and large scale text analysis techniques in low resource settings, especially in biomedical, life sciences and healthcare technologies. He enjoys playing soccer, water sports and traveling with his family.
 Garin Kessler is a Senior Data Science Manager at Amazon Web Services, where he leads teams of data scientists and application architects to deliver bespoke machine learning applications for customers. Outside of AWS, he lectures on machine learning and neural language models at Georgetown. When not working, he enjoys listening to (and making) music of questionable quality with friends and family.
Garin Kessler is a Senior Data Science Manager at Amazon Web Services, where he leads teams of data scientists and application architects to deliver bespoke machine learning applications for customers. Outside of AWS, he lectures on machine learning and neural language models at Georgetown. When not working, he enjoys listening to (and making) music of questionable quality with friends and family.
 Dr. Taha Kass-Hout is Director of Machine Learning and Chief Medical Officer at Amazon Web Services, and leads our Health AI strategy and efforts, including Amazon Comprehend Medical and Amazon HealthLake. Taha is also working with teams at Amazon responsible for developing the science, technology, and scale for COVID-19 lab testing. A physician and bioinformatician, Taha served two terms under President Obama, including the first Chief Health Informatics officer at the FDA. During this time as a public servant, he pioneered the use of emerging technologies and cloud (CDC’s electronic disease surveillance), and established widely accessible global data sharing platforms, the openFDA, that enabled researchers and the public to search and analyze adverse event data, and precisionFDA (part of the Presidential Precision Medicine initiative).
Dr. Taha Kass-Hout is Director of Machine Learning and Chief Medical Officer at Amazon Web Services, and leads our Health AI strategy and efforts, including Amazon Comprehend Medical and Amazon HealthLake. Taha is also working with teams at Amazon responsible for developing the science, technology, and scale for COVID-19 lab testing. A physician and bioinformatician, Taha served two terms under President Obama, including the first Chief Health Informatics officer at the FDA. During this time as a public servant, he pioneered the use of emerging technologies and cloud (CDC’s electronic disease surveillance), and established widely accessible global data sharing platforms, the openFDA, that enabled researchers and the public to search and analyze adverse event data, and precisionFDA (part of the Presidential Precision Medicine initiative).
