Generative AI models have been experiencing rapid growth in recent months due to its impressive capabilities in creating realistic text, images, code, and audio. Among these models, Stable Diffusion models stand out for their unique strength in creating high-quality images based on text prompts. Stable Diffusion can generate a wide variety of high-quality images, including realistic portraits, landscapes, and even abstract art. And, like other generative AI models, Stable Diffusion models require powerful computing to provide low-latency inference.
In this post, we show how you can run Stable Diffusion models and achieve high performance at the lowest cost in Amazon Elastic Compute Cloud (Amazon EC2) using Amazon EC2 Inf2 instances powered by AWS Inferentia2. We look at the architecture of a Stable Diffusion model and walk through the steps of compiling a Stable Diffusion model using AWS Neuron and deploying it to an Inf2 instance. We also discuss the optimizations that the Neuron SDK automatically makes to improve performance. You can run both Stable Diffusion 2.1 and 1.5 versions on AWS Inferentia2 cost-effectively. Lastly, we show how you can deploy a Stable Diffusion model to an Inf2 instance with Amazon SageMaker.
The Stable Diffusion 2.1 model size in floating point 32 (FP32) is 5 GB and 2.5 GB in bfoat16 (BF16). A single inf2.xlarge instance has one AWS Inferentia2 accelerator with 32 GB of HBM memory. The Stable Diffusion 2.1 model can fit on a single inf2.xlarge instance. Stable Diffusion is a text-to-image model that you can use to create images of different styles and content simply by providing a text prompt as an input. To learn more about the Stable Diffusion model architecture, refer to Create high-quality images with Stable Diffusion models and deploy them cost-efficiently with Amazon SageMaker.
How the Neuron SDK optimizes Stable Diffusion performance
Before we can deploy the Stable Diffusion 2.1 model on AWS Inferentia2 instances, we need to compile the model components using the Neuron SDK. The Neuron SDK, which includes a deep learning compiler, runtime, and tools, compiles and automatically optimizes deep learning models so they can run efficiently on Inf2 instances and extract full performance of the AWS Inferentia2 accelerator. We have examples available for Stable Diffusion 2.1 model on the GitHub repo. This notebook presents an end-to-end example of how to compile a Stable Diffusion model, save the compiled Neuron models, and load it into the runtime for inference.
We use StableDiffusionPipeline from the Hugging Face diffusers library to load and compile the model. We then compile all the components of the model for Neuron using torch_neuronx.trace() and save the optimized model as TorchScript. Compilation processes can be quite memory-intensive, requiring a significant amount of RAM. To circumvent this, before tracing each model, we create a deepcopy of the part of the pipeline that’s being traced. Following this, we delete the pipeline object from memory using del pipe. This technique is particularly useful when compiling on instances with low RAM.
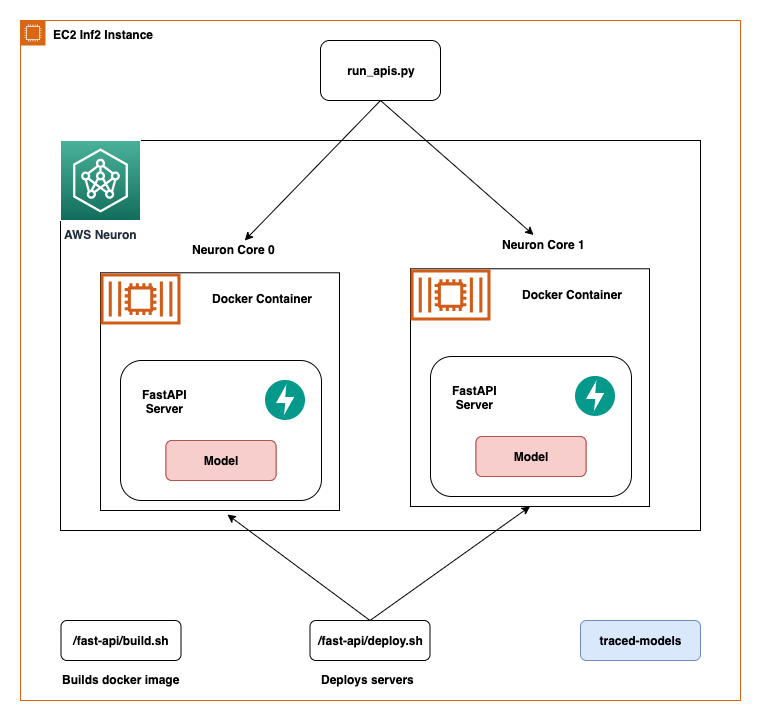
Additionally, we also perform optimizations to the Stable Diffusion models. UNet holds the most computationally intensive aspect of the inference. The UNet component operates on input tensors that have a batch size of two, generating a corresponding output tensor also with a batch size of two, to produce a single image. The elements within these batches are entirely independent of each other. We can take advantage of this behavior to get optimal latency by running one batch on each Neuron core. We compile the UNet for one batch (by using input tensors with one batch), then use the torch_neuronx.DataParallel API to load this single batch model onto each core. The output of this API is a seamless two-batch module: we can pass to the UNet the inputs of two batches, and a two-batch output is returned, but internally, the two single-batch models are running on the two Neuron cores. This strategy optimizes resource utilization and reduces latency.
Compile and deploy a Stable Diffusion model on an Inf2 EC2 instance
To compile and deploy the Stable Diffusion model on an Inf2 EC2 instance, sign to the AWS Management Console and create an inf2.8xlarge instance. Note that an inf2.8xlarge instance is required only for the compilation of the model because compilation requires a higher host memory. The Stable Diffusion model can be hosted on an inf2.xlarge instance. You can find the latest AMI with Neuron libraries using the following AWS Command Line Interface (AWS CLI) command:
For this example, we created an EC2 instance using the Deep Learning AMI Neuron PyTorch 1.13 (Ubuntu 20.04). You can then create a JupyterLab lab environment by connecting to the instance and running the following steps:
A notebook with all the steps for compiling and hosting the model is located on GitHub.
Let’s look at the compilation steps for one of the text encoder blocks. Other blocks that are part of the Stable Diffusion pipeline can be compiled similarly.
The first step is to load the pre-trained model from Hugging Face. The StableDiffusionPipeline.from_pretrained method loads the pre-trained model into our pipeline object, pipe. We then create a deepcopy of the text encoder from our pipeline, effectively cloning it. The del pipe command is then used to delete the original pipeline object, freeing up the memory that was consumed by it. Here, we are quantizing the model to BF16 weights:
This step involves wrapping our text encoder with the NeuronTextEncoder wrapper. The output of a compiled text encoder module will be of dict. We convert it to a list type using this wrapper:
We initialize PyTorch tensor emb with some values. The emb tensor is used as example input for the torch_neuronx.trace function. This function traces our text encoder and compiles it into a format optimized for Neuron. The directory path for the compiled model is constructed by joining COMPILER_WORKDIR_ROOT with the subdirectory text_encoder:
The compiled text encoder is saved using torch.jit.save. It’s stored under the file name model.pt in the text_encoder directory of our compiler’s workspace:
The notebook includes similar steps to compile other components of the model: UNet, VAE decoder, and VAE post_quant_conv. After you have compiled all the models, you can load and run the model following these steps:
- Define the paths for the compiled models.
- Load a pre-trained
StableDiffusionPipelinemodel, with its configuration specified to use the bfloat16 data type. - Load the UNet model onto two Neuron cores using the
torch_neuronx.DataParallelAPI. This allows data parallel inference to be performed, which can significantly speed up model performance. - Load the remaining parts of the model (
text_encoder,decoder, andpost_quant_conv) onto a single Neuron core.
You can then run the pipeline by providing input text as prompts. The following are some pictures generated by the model for the prompts:
- Portrait of renaud sechan, pen and ink, intricate line drawings, by craig mullins, ruan jia, kentaro miura, greg rutkowski, loundraw

- Portrait of old coal miner in 19th century, beautiful painting, with highly detailed face painting by greg rutkowski

- A castle in the middle of a forest

Host Stable Diffusion 2.1 on AWS Inferentia2 and SageMaker
Hosting Stable Diffusion models with SageMaker also requires compilation with the Neuron SDK. You can complete the compilation ahead of time or during runtime using Large Model Inference (LMI) containers. Compilation ahead of time allows for faster model loading times and is the preferred option.
SageMaker LMI containers provide two ways to deploy the model:
- A no-code option where we just provide a
serving.propertiesfile with the required configurations - Bring your own inference script
We look at both solutions and go over the configurations and the inference script (model.py). In this post, we demonstrate the deployment using a pre-compiled model stored in an Amazon Simple Storage Service (Amazon S3) bucket. You can use this pre-compiled model for your deployments.
Configure the model with a provided script
In this section, we show how to configure the LMI container to host the Stable Diffusion models. The SD2.1 notebook available on GitHub. The first step is to create the model configuration package per the following directory structure. Our aim is to use the minimal model configurations needed to host the model. The directory structure needed is as follows:
Next, we create the serving.properties file with the following parameters:
The parameters specify the following:
- option.model_id – The LMI containers use s5cmd to load the model from the S3 location and therefore we need to specify the location of where our compiled weights are.
- option.entryPoint – To use the built-in handlers, we specify the transformers-neuronx class. If you have a custom inference script, you need to provide that instead.
- option.dtype – This specifies to load the weights in a specific size. For this post, we use BF16, which further reduces our memory requirements vs. FP32 and lowers our latency due to that.
- option.tensor_parallel_degree – This parameter specifies the number of accelerators we use for this model. The AWS Inferentia2 chip accelerator has two Neuron cores and so specifying a value of 2 means we use one accelerator (two cores). This means we can now create multiple workers to increase the throughput of the endpoint.
- option.engine – This is set to Python to indicate we will not be using other compilers like DeepSpeed or Faster Transformer for this hosting.
Bring your own script
If you want to bring your own custom inference script, you need to remove the option.entryPoint from serving.properties. The LMI container in that case will look for a model.py file in the same location as the serving.properties and use that to run the inferencing.
Create your own inference script (model.py)
Creating your own inference script is relatively straightforward using the LMI container. The container requires your model.py file to have an implementation of the following method:
Let’s examine some of the critical areas of the attached notebook, which demonstrates the bring your own script function.
Replace the cross_attention module with the optimized version:
These are the names of the compiled weights file we used when creating the compilations. Feel free to change the file names, but make sure your weights file names match what you specify here.
Then we need to load them using the Neuron SDK and set these in the actual model weights. When loading the UNet optimized weights, note we are also specifying the number of Neuron cores we need to load these onto. Here, we load to a single accelerator with two cores:
Running the inference with a prompt invokes the pipe object to generate an image.
Create the SageMaker endpoint
We use Boto3 APIs to create a SageMaker endpoint. Complete the following steps:
- Create the tarball with just the serving and the optional
model.pyfiles and upload it to Amazon S3. - Create the model using the image container and the model tarball uploaded earlier.
- Create the endpoint config using the following key parameters:
- Use an
ml.inf2.xlargeinstance. - Set
ContainerStartupHealthCheckTimeoutInSecondsto 240 to ensure the health check starts after the model is deployed. - Set
VolumeInGBto a larger value so it can be used for loading the model weights that are 32 GB in size.
- Use an
Create a SageMaker model
After you create the model.tar.gz file and upload it to Amazon S3, we need to create a SageMaker model. We use the LMI container and the model artifact from the previous step to create the SageMaker model. SageMaker allows us to customize and inject various environment variables. For this workflow, we can leave everything as default. See the following code:
Create the model object, which essentially creates a lockdown container that is loaded onto the instance and used for inferencing:
Create a SageMaker endpoint
In this demo, we use an ml.inf2.xlarge instance. We need to set the VolumeSizeInGB parameters to provide the necessary disk space to load the model and the weights. This parameter is applicable to instances supporting the Amazon Elastic Block Store (Amazon EBS) volume attachment. We can leave the model download timeout and container startup health check to a higher value, which will give adequate time for the container to pull the weights from Amazon S3 and load into the AWS Inferentia2 accelerators. For more details, refer to CreateEndpointConfig.
Lastly, we create a SageMaker endpoint:
Invoke the model endpoint
This is a generative model, so we pass in the prompt that the model uses to generate the image. The payload is of the type JSON:
Benchmarking the Stable Diffusion model on Inf2
We ran a few tests to benchmark the Stable Diffusion model with BF 16 data type on Inf2, and we are able to derive latency numbers that rival or exceed some of the other accelerators for Stable Diffusion. This, coupled with the lower cost of AWS Inferentia2 chips, makes this an extremely valuable proposition.
The following numbers are from the Stable Diffusion model deployed on an inf2.xl instance. For more information about costs, refer to Amazon EC2 Inf2 Instances.
| Model | Resolution | Data type | Iterations | P95 Latency (ms) | Inf2.xl On-Demand cost per hour | Inf2.xl (Cost per image) |
| Stable Diffusion 1.5 | 512×512 | bf16 | 50 | 2,427.4 | $0.76 | $0.0005125 |
| Stable Diffusion 1.5 | 768×768 | bf16 | 50 | 8,235.9 | $0.76 | $0.0017387 |
| Stable Diffusion 1.5 | 512×512 | bf16 | 30 | 1,456.5 | $0.76 | $0.0003075 |
| Stable Diffusion 1.5 | 768×768 | bf16 | 30 | 4,941.6 | $0.76 | $0.0010432 |
| Stable Diffusion 2.1 | 512×512 | bf16 | 50 | 1,976.9 | $0.76 | $0.0004174 |
| Stable Diffusion 2.1 | 768×768 | bf16 | 50 | 6,836.3 | $0.76 | $0.0014432 |
| Stable Diffusion 2.1 | 512×512 | bf16 | 30 | 1,186.2 | $0.76 | $0.0002504 |
| Stable Diffusion 2.1 | 768×768 | bf16 | 30 | 4,101.8 | $0.76 | $0.0008659 |
Conclusion
In this post, we dove deep into the compilation, optimization, and deployment of the Stable Diffusion 2.1 model using Inf2 instances. We also demonstrated deployment of Stable Diffusion models using SageMaker. Inf2 instances also deliver great price performance for Stable Diffusion 1.5. To learn more about why Inf2 instances are great for generative AI and large language models, refer to Amazon EC2 Inf2 Instances for Low-Cost, High-Performance Generative AI Inference are Now Generally Available. For performance details, refer to Inf2 Performance. Check out additional examples on the GitHub repo.
Special thanks to Matthew Mcclain, Beni Hegedus, Kamran Khan, Shruti Koparkar, and Qing Lan for reviewing and providing valuable inputs.
About the Authors
 Vivek Gangasani is a Senior Machine Learning Solutions Architect at Amazon Web Services. He works with machine learning startups to build and deploy AI/ML applications on AWS. He is currently focused on delivering solutions for MLOps, ML inference, and low-code ML. He has worked on projects in different domains, including natural language processing and computer vision.
Vivek Gangasani is a Senior Machine Learning Solutions Architect at Amazon Web Services. He works with machine learning startups to build and deploy AI/ML applications on AWS. He is currently focused on delivering solutions for MLOps, ML inference, and low-code ML. He has worked on projects in different domains, including natural language processing and computer vision.
 K.C. Tung is a Senior Solution Architect in AWS Annapurna Labs. He specializes in large deep learning model training and deployment at scale in cloud. He has a Ph.D. in molecular biophysics from the University of Texas Southwestern Medical Center in Dallas. He has spoken at AWS Summits and AWS Reinvent. Today he helps customers to train and deploy large PyTorch and TensorFlow models in AWS cloud. He is the author of two books: Learn TensorFlow Enterprise and TensorFlow 2 Pocket Reference.
K.C. Tung is a Senior Solution Architect in AWS Annapurna Labs. He specializes in large deep learning model training and deployment at scale in cloud. He has a Ph.D. in molecular biophysics from the University of Texas Southwestern Medical Center in Dallas. He has spoken at AWS Summits and AWS Reinvent. Today he helps customers to train and deploy large PyTorch and TensorFlow models in AWS cloud. He is the author of two books: Learn TensorFlow Enterprise and TensorFlow 2 Pocket Reference.
 Rupinder Grewal is a Sr Ai/ML Specialist Solutions Architect with AWS. He currently focuses on serving of models and MLOps on SageMaker. Prior to this role he has worked as Machine Learning Engineer building and hosting models. Outside of work he enjoys playing tennis and biking on mountain trails.
Rupinder Grewal is a Sr Ai/ML Specialist Solutions Architect with AWS. He currently focuses on serving of models and MLOps on SageMaker. Prior to this role he has worked as Machine Learning Engineer building and hosting models. Outside of work he enjoys playing tennis and biking on mountain trails.


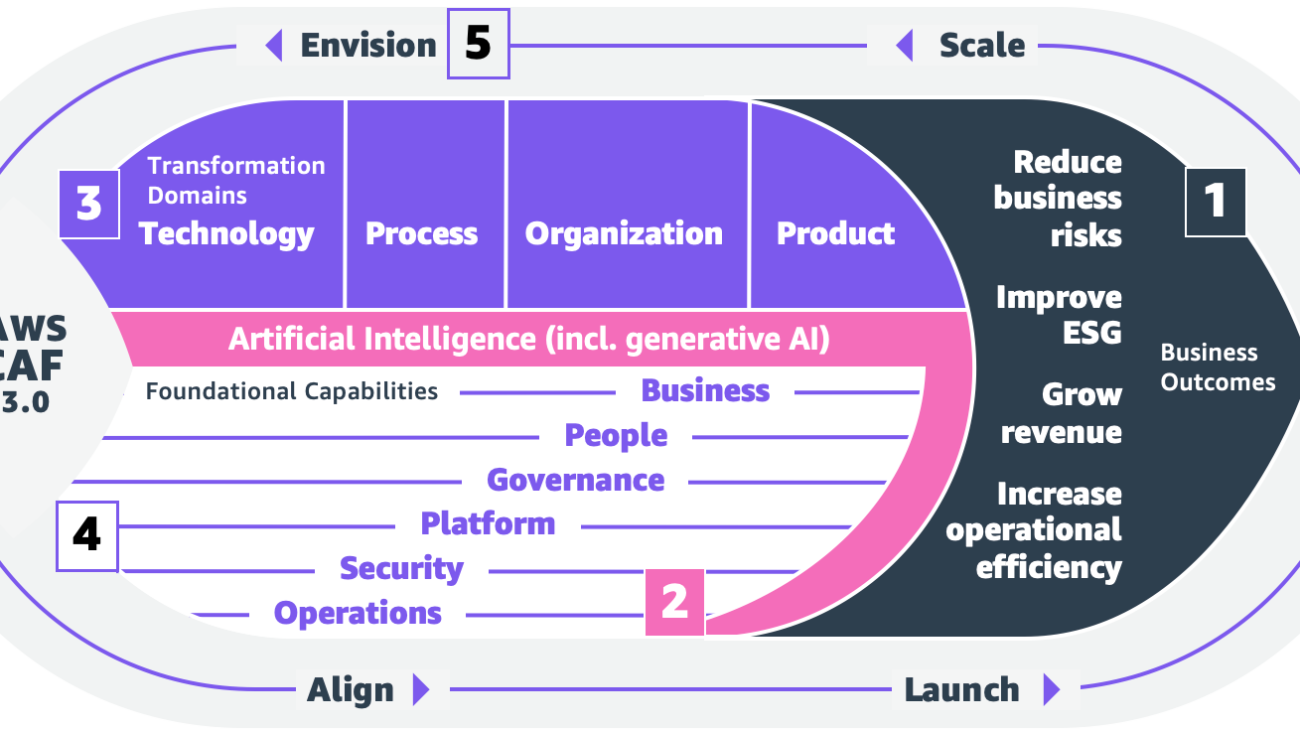
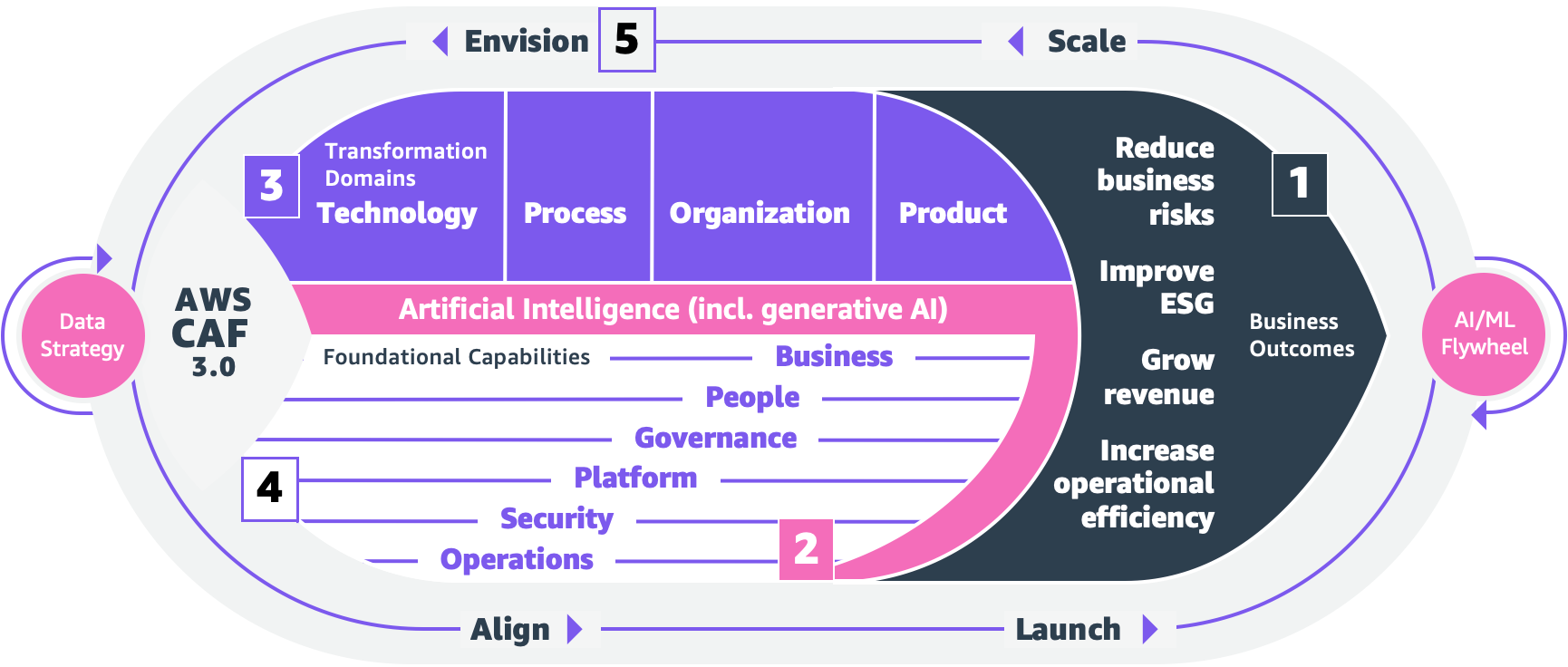

 Caleb Wilkinson has more than a decade of experience building AI solutions. As a Senior Machine Learning Strategist at AWS, Caleb pioneers innovative applications of AI that push the boundaries of possibility and helps organizations benefit responsibly from artificial intelligence. He is the co-author of CAF-AI.
Caleb Wilkinson has more than a decade of experience building AI solutions. As a Senior Machine Learning Strategist at AWS, Caleb pioneers innovative applications of AI that push the boundaries of possibility and helps organizations benefit responsibly from artificial intelligence. He is the co-author of CAF-AI. Alexander Wöhlke has a decade of experience in AI and ML. He is Senior Machine Learning Strategist and Technical Product Manager at the AWS Generative AI Innovation Center. He works with large organizations on their AI-Strategy and helps them take calculated risks at the forefront of technological development. He is the co-author of CAF-AI.
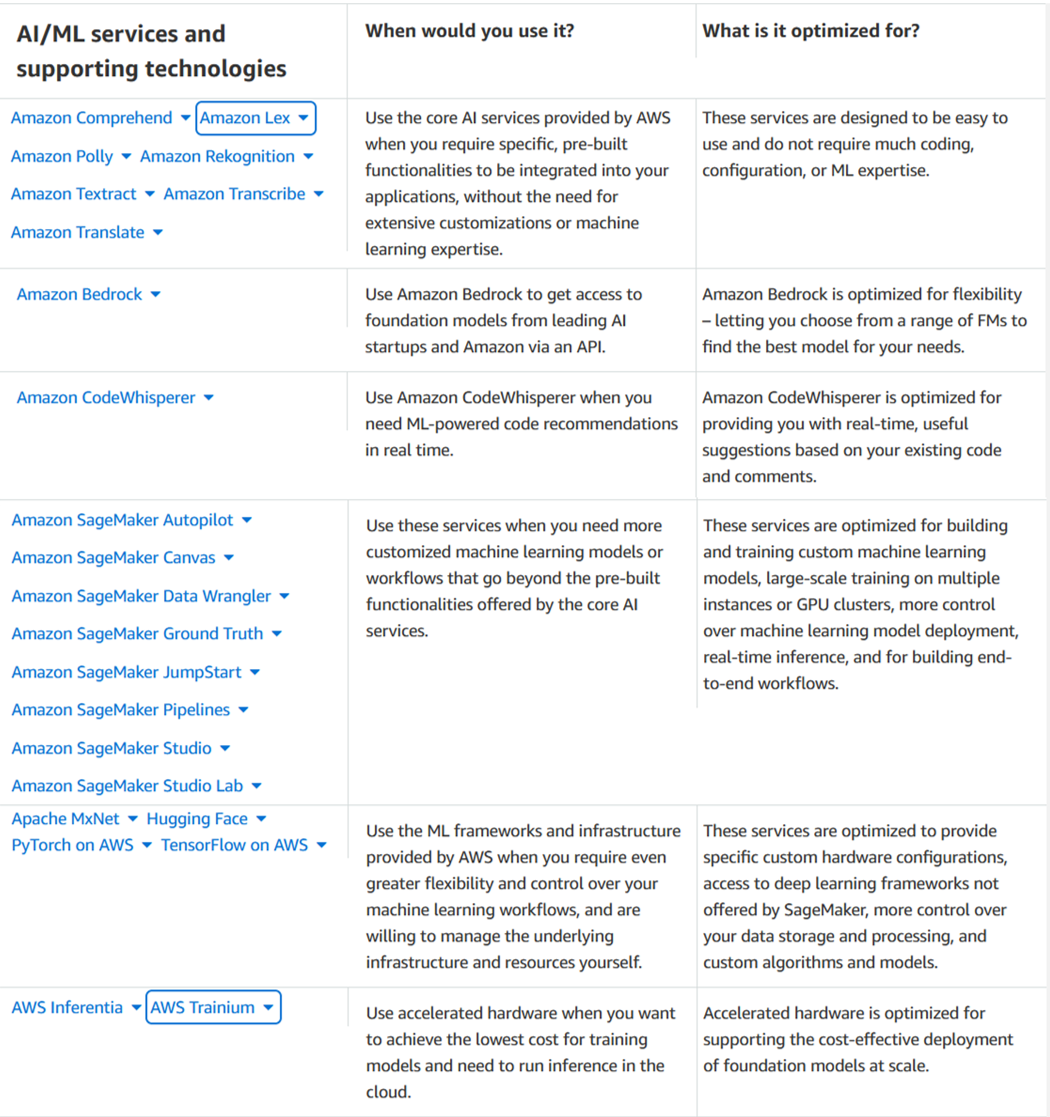
Alexander Wöhlke has a decade of experience in AI and ML. He is Senior Machine Learning Strategist and Technical Product Manager at the AWS Generative AI Innovation Center. He works with large organizations on their AI-Strategy and helps them take calculated risks at the forefront of technological development. He is the co-author of CAF-AI. Geof Wheelwright manages the AWS decision content team, which writes and develops the growing collection of decision guides on the AWS Getting Started Resource Center. His team created the Choosing an AWS machine learning decision guide. He has enjoyed working with AI and its ancestors since first being introduced to simple, text-based Apple II
Geof Wheelwright manages the AWS decision content team, which writes and develops the growing collection of decision guides on the AWS Getting Started Resource Center. His team created the Choosing an AWS machine learning decision guide. He has enjoyed working with AI and its ancestors since first being introduced to simple, text-based Apple II 

















 Mehran Nikoo is a Senior Solutions Architect at AWS, working with Digital Native businesses in the UK and helping them achieve their goals. Passionate about applying his software engineering experience to machine learning, he specializes in end-to-end machine learning and MLOps practices.
Mehran Nikoo is a Senior Solutions Architect at AWS, working with Digital Native businesses in the UK and helping them achieve their goals. Passionate about applying his software engineering experience to machine learning, he specializes in end-to-end machine learning and MLOps practices.




 Guang Yang is a senior applied scientist at the AWS Generative AI Innovation Center where he works with customers across various verticals and applies creative problem solving to generate value for customers with state-of-the-art ML/AI solutions.
Guang Yang is a senior applied scientist at the AWS Generative AI Innovation Center where he works with customers across various verticals and applies creative problem solving to generate value for customers with state-of-the-art ML/AI solutions.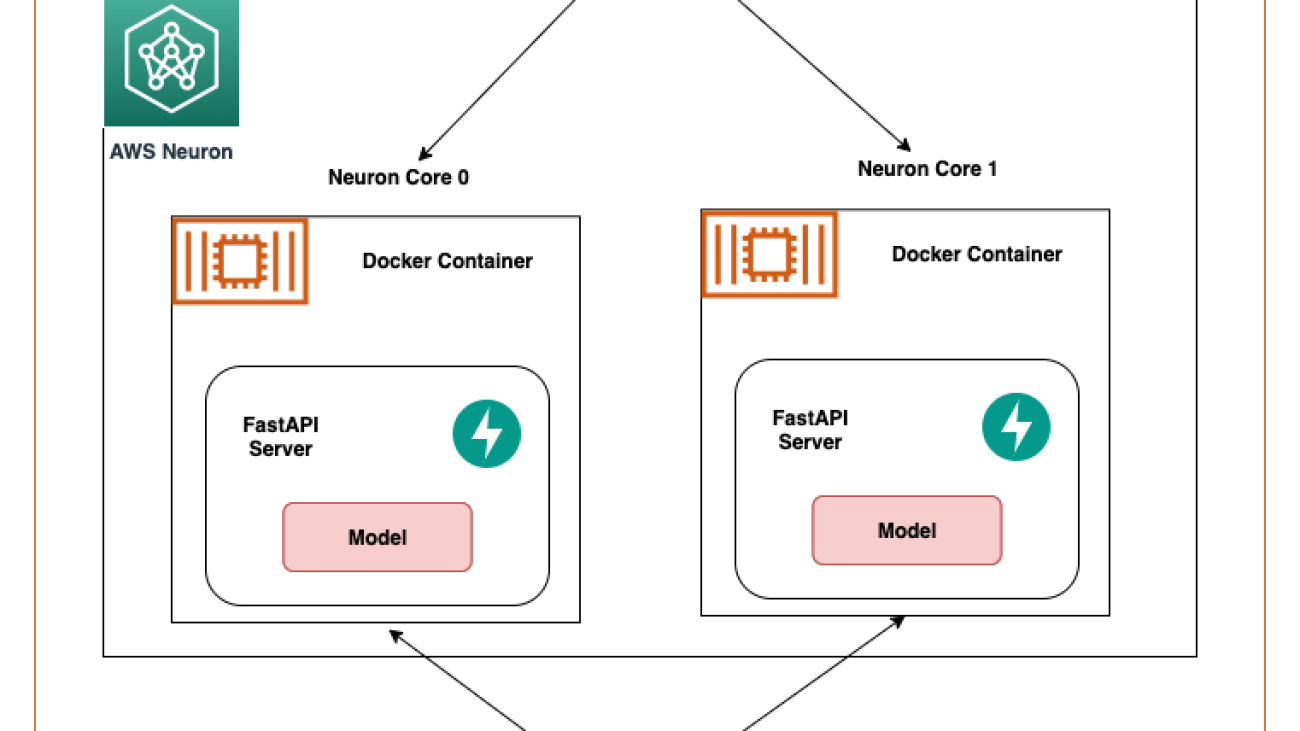
 Ankur Srivastava is a Sr. Solutions Architect in the ML Frameworks Team. He focuses on helping customers with self-managed distributed training and inference at scale on AWS. His experience includes industrial predictive maintenance, digital twins, probabilistic design optimization and has completed his doctoral studies from Mechanical Engineering at Rice University and post-doctoral research from Massachusetts Institute of Technology.
Ankur Srivastava is a Sr. Solutions Architect in the ML Frameworks Team. He focuses on helping customers with self-managed distributed training and inference at scale on AWS. His experience includes industrial predictive maintenance, digital twins, probabilistic design optimization and has completed his doctoral studies from Mechanical Engineering at Rice University and post-doctoral research from Massachusetts Institute of Technology. Pronoy Chopra is a Senior Solutions Architect with the Startups Generative AI team at AWS. He specializes in architecting and developing IoT and Machine Learning solutions. He has co-founded two startups in the past and enjoys being hands-on with projects in the IoT, AI/ML and Serverless domain.
Pronoy Chopra is a Senior Solutions Architect with the Startups Generative AI team at AWS. He specializes in architecting and developing IoT and Machine Learning solutions. He has co-founded two startups in the past and enjoys being hands-on with projects in the IoT, AI/ML and Serverless domain.