Drug discovery is like searching for the right jigsaw tile — in a puzzle box with 1060 molecular-size pieces. AI and HPC tools help researchers more quickly narrow down the options, like picking out a subset of correctly shaped and colored puzzle pieces to experiment with.
An effective small-molecule drug will bind to a target enzyme, receptor or other critical protein along the disease pathway. Like the perfect puzzle piece, a successful drug will be the ideal fit, possessing the right shape, flexibility and interaction energy to attach to its target.
But it’s not enough just to interact strongly with the target. An effective therapeutic must modify the function of the protein in just the right way, and also possess favorable absorption, distribution, metabolism, excretion and toxicity properties — creating a complex optimization problem for scientists.
Researchers worldwide are racing to find effective vaccine and drug candidates to inhibit infection with and replication of SARS-CoV-2, the virus that causes COVID-19. Using NVIDIA GPUs, they’re accelerating this lengthy discovery process — whether for structure-based drug design, molecular docking, generative AI models, virtual screening or high-throughput screening.
Identifying Protein Targets with Genomics
To develop an effective drug, researchers have to know where to start. A disease pathway — a chain of signals between molecules that trigger different cell functions — may involve thousands of interacting proteins. Genomic analyses can provide invaluable insights for researchers, helping them identify promising proteins to target with a specific drug.
With the NVIDIA Clara Parabricks genome analysis toolkit, researchers can sequence and analyze genomes up to 50x faster. Given the unprecedented spread of the COVID pandemic, getting results in hours versus days can have an extraordinary impact on understanding the virus and developing treatments.
To date, hundreds of institutions, including hospitals, universities and supercomputing centers, in 88 countries have downloaded the software to accelerate their work — to sequence the viral genome itself, as well as to sequence the DNA of COVID patients and investigate why some are more severely affected by the virus than others.
Another method, cryo-EM, uses electron microscopes to directly observe flash-frozen proteins — and can harness GPUs to shorten processing time for the complex, massive datasets involved.
Using CryoSPARC, a GPU-accelerated software built by Toronto startup Structura Biotechnology, researchers at the National Institutes of Health and the University of Texas at Austin created the first 3D, atomic-scale map of the coronavirus, providing a detailed view into the virus’ spike proteins, a key target for vaccines, therapeutic antibodies and diagnostics.
GPU-Accelerated Compound Screening
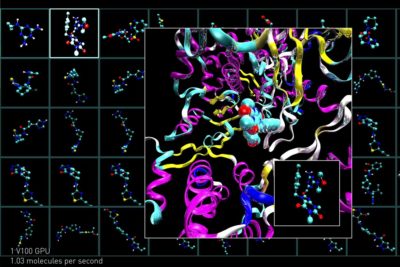 Once a target protein has been identified, researchers search for candidate compounds that have the right properties to bind with it. To evaluate how effective drug candidates will be, researchers can screen drug candidates virtually, as well as in real-world labs.
Once a target protein has been identified, researchers search for candidate compounds that have the right properties to bind with it. To evaluate how effective drug candidates will be, researchers can screen drug candidates virtually, as well as in real-world labs.
New York-based Schrödinger creates drug discovery software that can model the properties of potential drug molecules. Used by the world’s biggest biopharma companies, the Schrödinger platform allows its users to determine the binding affinity of a candidate molecule on NVIDIA Tensor Core GPUs in under an hour and with just a few dollars of compute cost — instead of many days and thousands of dollars using traditional methods.
Generative AI Models for Drug Discovery
Rather than evaluating a dataset of known drug candidates, a generative AI model starts from scratch. Tokyo-based startup Elix, Inc., a member of the NVIDIA Inception virtual accelerator program, uses generative models trained on NVIDIA DGX Station systems to come up with promising molecular structures. Some of the AI’s proposed molecules may be unstable or difficult to synthesize, so additional neural networks are used to determine the feasibility for these candidates to be tested in the lab.
With DGX Station, Elix achieves up to a 6x speedup on training the generative models, which would otherwise take a week or more to converge, or to reach the lowest possible error rate.
Molecular Docking for COVID-19 Research
With the inconceivable size of the chemical space, researchers couldn’t possibly test every possible molecule to figure out which will be effective to combat a specific disease. But based on what’s known about the target protein, GPU-accelerated molecular dynamics applications can be used to approximate molecular behavior and simulate target proteins at the atomic level.
Software like AutoDock-GPU, developed by the Center for Computational Structural Biology at the Scripps Research Institute, enables researchers to calculate the interaction energy between a candidate molecule and the protein target. Known as molecular docking, this computationally complex process simulates millions of different configurations to find the most favorable arrangement of each molecule for binding. Using the more than 27,000 NVIDIA GPUs on Oak Ridge National Laboratory’s Summit supercomputer, scientists were able to screen 1 billion drug candidates for COVID-19 in just 12 hours. Even using a single NVIDIA GPU provides more than 230x speedup over using a single CPU.

In Illinois, Argonne National Laboratory is accelerating COVID-19 research using an NVIDIA A100 GPU-powered system based on the DGX SuperPOD reference architecture. Argonne researchers are combining AI and advanced molecular modelling methods to perform accelerated simulations of the viral proteins, and to screen billions of potential drug candidates, determining the most promising molecules to pursue for clinical trials.
Accelerating Biological Image Analysis
The drug discovery process involves significant high-throughput lab experiments as well. Phenotypic screening is one method of testing, in which a diseased cell is exposed to a candidate drug. With microscopes, researchers can observe and record subtle changes in the cell to determine if it starts to more closely resemble a healthy cell. Using AI to automate the process, thousands of possible drugs can be screened.
Digital biology company Recursion, based in Salt Lake City, uses AI and NVIDIA GPUs to observe these subtle changes in cell images, analyzing terabytes of data each week. The company has released an open-source COVID dataset, sharing human cellular morphological data with researchers working to create therapies for the virus.
Future Directions in AI for Drug Discovery
As AI and accelerated computing continue to accelerate genomics and drug discovery pipelines, precision medicine — personalizing individual patients’ treatment plans based on insights about their genome and their phenotype — will become more attainable.
Increasingly powerful NLP models will be applied to organize and understand massive datasets of scientific literature, helping connect the dots between independent investigations. Generative models will learn the fundamental equations of quantum mechanics and be able to suggest the optimal molecular therapy for a given target.
To learn more about how NVIDIA GPUs are being used to accelerate drug discovery, check out talks by Schrödinger, Oak Ridge National Laboratory and Atomwise at the GPU Technology Conference next week.
For more on how AI and GPUs are advancing COVID research, read our blog stories and visit the COVID-19 research hub.
Subscribe to NVIDIA healthcare news here.
The post Drug Discovery in the Age of COVID-19 appeared first on The Official NVIDIA Blog.
