A guest post by Jeffrey Ly, CEO & Joanna Ashby, CMO of mPOD, Inc.
mPOD is a NIH-funded pre-seed startup headquartered out of Johnson & Johnson’s Innovation (JLABS) in New York City. In this article, we’d like to share with you a hardware device we have developed independently at mPOD leveraging TensorFlow Lite Micro (TFLM) as a core technology, called DxTrack.
mPOD DxTrack leverages TFLM and low cost hardware to enable accurate, rapid and objective interpretation of currently available lateral flow assays (LFAs) in less than 10 seconds. LFAs serve as diagnostic tools because they are low-cost and simple to use without specialized skills or equipment. Most recently popularized by COVID-19 rapid antigen tests, LFAs are also used extensively testing for pregnancy, disease tracking, STDs, food intolerances, and therapeutic drugs along with an extensive array of biomarkers totaling billions of tests sold each year. The mPOD Dxtrack is applicable to use with any type of visually read lateral flow assay, demonstrating a healthcare use case for TFLM that can directly impact our everyday lives.
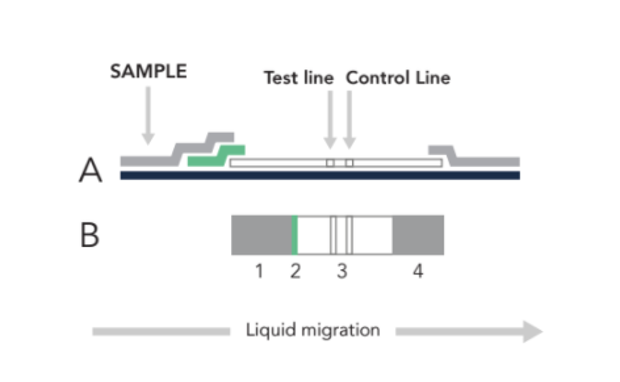
The LFA begins with a sample (nasal swab, saliva, urine, blood, etc) loaded at (1) in the figure below. Once the sample has flowed to the green conjugate zone (2), it is labeled with a signaling moiety. Through capillary action, the sample will continue flowing until it is immobilized at (3), with these LFA tests, two lines indicate a positive result, one line indicates a negative result.
 |
| Figure 1. Side (A) & Top (B) view of a lateral flow assay (LFA) sample where at (1) the sample (nasal swab, saliva, urine, blood, etc) is loaded before flowing to the green zone (2), where the target is labeled with a signaling moiety. Through capillary action, the sample will continue flowing until it is immobilized at (3) to form the test line. Excess material is absorbed at (4). |
 |
| Figure 2. These are the 3 possible classes results for a lateral flow assay (LFA) test. |
 |
| Figure 3. This is a diagram NOWDiagnostics ADEXUSDx lateral flow assay (LFA) designed to collect and run saliva sample in point-of-care (POC) and over-the-counter (OTC) settings. |
When used correctly, these tests are very effective; however self-testing presents challenges for the lay user to interpret. Significant variability is present between devices, making it difficult to tell if the test line you see is negative …or a faint positive?
 |
| Figure 4. A visualization of how the TinyML model on the mPOD DxTrack break interprets and classifies different lateral flow assay (LFA) results. |
To address this challenge, we developed mPOD DxTrack, an over-the-counter (OTC) LFA reader that improves the utility of lateral flow assays by enabling rapid and objective readings with a simple, under $5 (Cost-of-Goods) globally-deployable device. The mPOD DxTrack aims to read lateral flow assay tests using ML to accomplish two goals: 1) enable rapid and objective readings of LFAs and 2) streamline digital reporting. Critically, TinyML allows for the software on the mPOD DxTrack to be deployed on low-cost (less-than $5) hardware that can be widely distributed – which is difficult with existing LFA readers which rely on high-cost/high complexity hardware that cost hundreds to thousands of dollars per unit. Ultimately, we believe that TinyML will enable the mPOD DxTrack to catch missed positive test results by removing human bias and increasing confidence in lateral flow device testing, reducing user error, and increasing overall result accuracy.
 |
| Figure 5. Assembly view of the mPOD DxTrack with lateral flow assay (LFA) cassette. |
Technical Dive
Key Considerations
- Achieving high accuracy 99% overall accuracy, (99% sensitivity, 99% specificity) for model performance when interpreting live-run LFA strips.
- Ensuring the model can maintain that level of performance while fitting the hardware constraints.
Model size constraints for TinyML
Deployment of the DxTrack TinyML model on the Pico4ML Dev kit is constrained by 2 pieces of hardware: Flash memory and SRAM. The Pico4ML Dev kit has 2MB of flash memory to host the .uf2 file and 264kb of SRAM that accommodate the intermediate arrays (among other things) of the model. Ensuring the model size stays within these bounds is critical because while the code can successfully compile, run on the host machine and even successfully flash on the Pico4Ml Dev Kit, it will hang during set-up and not execute the main loop.
Rather than guess and check the size of intermediate arrays (a process we initially took with little reproducible success), we ended up developing a workflow that enabled us to quantify the model’s arena size by first using the interpreter function. See below, where this function was called during setup:
TfLiteStatus setup_status = ScreenInit(error_reporter);
if (setup_status != kTfLiteOk){
while(1){TF_LITE_REPORT_ERROR(error_reporter, "Set up failedn");};
}
arena_size = interpreter->arena_used_bytes();
printf("Arena_Size Used: %zu n", arena_size);When printed out, this is what the value from the interpreter function should look during Pico4ML Dev kit boot-up:
DEV_Module_Init OK
Arena_Size Used: 93500
sd_spi_go_low_frequency: Actual frequency: 122070
V2-Version Card
R3/R7: 0x1aa
R3/R7: 0xff8000
R3/R7: 0xc0ff8000
Card Initialized: High Capacity Card
SD card initialized
SDHC/SDXC Card: hc_c_size: 15237
Sectors: 15603712
Capacity: 7619 MB
sd_spi_go_high_frequency: Actual frequency: 12500000
With this value available to us, we are then able to set the appropriate TensorArenaSize. As you can see from above, the model uses 93500 bytes of SRAM. By setting the TensorArenaSize to just above that amount 99×1024 = 101376 bytes, we are able to allocate enough memory to host the model without going over the hardware limits (which also causes the Pico4ML Dev Kit to freeze).
// An area of memory to use for input, output, and intermediate arrays.
constexpr int kTensorArenaSize = 99* 1024; // 136 * 1024; //81 * 1024;
static uint8_t tensor_arena[kTensorArenaSize];Transforming from Unquantized to Quantized Models
Now that we have a reproducible methodology to quantify and deploy the model onto the Pico4ML Dev Kit, our next challenge is ensuring that the model can achieve the accuracy we require while still fitting with the size constrained by the hardware. For reference, the mPOD DxTrack platform is designed to interpret a 96×96 image. In the original model design, we were able to achieve > 99.999% accuracy with our model, but the intermediate layer is 96x96x32 at fp32 which requires over 1 MB of memory – it would never fit on the Pico4ML Dev Kit’s 264KB of SRAM. In order to achieve the size requirement for the model, we needed to take the model from unquantized to quantized; our best option was to utilize full int8 quantization. In essence, instead of treating the tensor values as floating points (float32), we correlate those values to integers (int8). Unfortunately, this decreased the model size 4-fold, allowing it to fit onto the Pico4ML Dev Kit’s rounding error from fp32 to int8 compounded, resulting in dramatically reduced model performance.
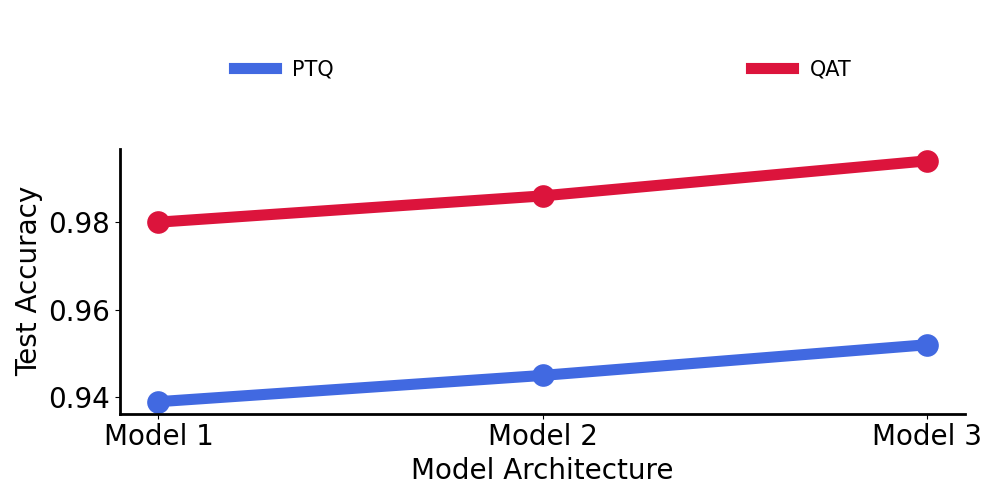
To combat this drop in model performance, we examined the effect of two different quantization strategies to improve performance: Post-training quantization (PTQ) and Quantization-aware training (QAT).
Below, we compare 3 different models to understand which quantization strategy is best. For reference:
- Model 1: 2-layer convolutional network
- Model 2: 3-layer convolutional network
- Model 3: 4-layer convolutional network
As we can see, Quantization-aware training (QAT) uniformly beats the post-training quantization (PTQ) method and it became part of our workflow moving forward.
What performance can we achieve now?
Tested across over 800 real-world test runs, the mPOD DxTrack can preliminary achieve an overall accuracy of 98.7%. This version of the model is currently being evaluated by our network of manufacturing partners who we work closely with. Currently we are assembling a unique dataset of images as part of a patient-focused data pipeline to learn from each manufacturing partnership and building bespoke models.
Our preliminary work has also helped us correlate model performance with appropriately large dataset size to achieve the performance high enough accuracy for our healthcare application. Per the figure attached, the model needs to be trained on a quality dataset of at least 15,000 images. Our commercial-ready target is likely to require datasets that are greater than 100,000 images.

To learn more about mPOD Inc, please visit our website at www.mpod.io. If you’re interested in learning more about TinyML, we recommend checking out this book and this course.


